Per S. Schmidt and Kristian S. Thygesen
The Journal of Physical Chemistry C Article ASAP
This database contains the adsorption energies of 200 reactions involving 8 different adsorbates on 25 different transition metal surfaces at full coverage as well as the surface energies for the same surfaces. Various DFT functionals have been employed and compared to the results from many-body perturbation theory within the random phase approximation.
The adsorption reactions are
\(\mathrm{H}_2\mathrm{O} + \mathrm{slab} \rightarrow \mathrm{OH/slab} + \frac{1}{2} \mathrm{H}_2\)
\(\mathrm{CH}_4 + \mathrm{slab} \rightarrow \mathrm{CH/slab} + \frac{3}{2} \mathrm{H}_2\)
\(\mathrm{NO} + \mathrm{slab} \rightarrow \mathrm{NO/slab}\)
\(\mathrm{CO} + \mathrm{slab} \rightarrow \mathrm{CO/slab}\)
\(\mathrm{N}_2 + \mathrm{slab} \rightarrow \mathrm{N_2/slab}\)
\(\frac{1}{2}\mathrm{N}_2 + \mathrm{slab} \rightarrow \mathrm{N/slab}\)
\(\frac{1}{2}\mathrm{O}_2 + \mathrm{slab} \rightarrow \mathrm{O/slab}\)
\(\frac{1}{2}\mathrm{H}_2 + \mathrm{slab} \rightarrow \mathrm{H/slab}\)
and the surfaces include 3d transition metals from Scandium (Sc) to Zink (Zn), 4d from Yttrium (Y) to Cadmium (Cd) excluding Technetium (Tc) and 5d from Hafnium (Hf) to Gold (Au).
Per S. Schmidt and Kristian S. Thygesen
The Journal of Physical Chemistry C Article ASAP
The data can be obtained from the files:
Download raw data adsorption.db and surfaces.db
And browsed online:
The adsorption energy is defined with respect to the adsorbate in its gas phase: \(E_{\text{ads}} = E_{\text{adsorbate@slab}} - (E_{\text{slab}} + E_{\text{adsorbate(g)}})\)
And the surface energy as: \(E_{\text{surf}} = \frac12 \bigg( E_{\text{slab}} - N_{\text{layers}} E_{\text{bulk}}\bigg)\), where the number of atomic layers in the slab is \(N_{\text{layers}} = 3\).
One example reaction that the adsorption energy is calculated for is:
\(\mathrm{CH}_4\mathrm{(g)} + \mathrm{Sc} \rightarrow \mathrm{CH/Sc} + \frac32 \mathrm{H}_2\mathrm{(g)}\)
In the database the adsorbate is then \(\mathrm{CH}\), the surface material is \(\mathrm{Sc}\), reference molecule 1 is \(\mathrm{CH}_4\) and reference molecule 2 is \(\mathrm{H}_2\). The adsorption energy is then \(E_{\text{ads}} = E_{\mathrm{CH/Sc}} -(E_{\mathrm{Sc}} + E_{\mathrm{CH}_4\mathrm{(g)}} - \frac32 E_{\mathrm{H}_2\mathrm{(g)}})\).
Database of adsorption energies:
key |
description |
unit |
|---|---|---|
BEEFvdW_ads |
BEEF-vdW adsorbate on slab |
eV |
BEEFvdW_adsorp |
Adsorption energy with BEEF-vdW |
eV |
BEEFvdW_mol |
BEEF-vdW 1st molecule |
eV |
BEEFvdW_mol2 |
BEEF-vdW 2nd molecule |
eV |
BEEFvdW_slab |
BEEF-vdW slab |
eV |
EXX_ads |
EXX adsorbate on slab |
eV |
EXX_adsorp |
Adsorption energy with EXX |
eV |
EXX_mol |
EXX 1st molecule |
eV |
EXX_mol2 |
EXX 2nd molecule |
eV |
EXX_slab |
EXX slab |
eV |
LDA_ads |
LDA adsorbate on slab |
eV |
LDA_adsorp |
Adsorption energy with LDA |
eV |
LDA_mol |
LDA 1st molecule |
eV |
LDA_mol2 |
LDA 2nd molecule |
eV |
LDA_slab |
LDA slab |
eV |
PBE_ads |
PBE adsorbate on slab |
eV |
PBE_adsorp |
Adsorption energy with PBE |
eV |
PBE_mol |
PBE 1st molecule |
eV |
PBE_mol2 |
PBE 2nd molecule |
eV |
PBE_slab |
PBE slab |
eV |
RPA_EXX_adsorp |
Adsorption energy with EXX+RPA |
eV |
RPA_ads_ecut300_k12 |
RPA adsorbate on slab, Ecut=300, k=12x12x1 |
eV |
RPA_ads_ecut300_k6 |
RPA adsorbate on slab, Ecut=300, k=6x6x1 |
eV |
RPA_ads_ecut400_k6 |
RPA adsorbate on slab, Ecut=400, k=6x6x1 |
eV |
RPA_ads_ecut500_k6 |
RPA adsorbate on slab, Ecut=500, k=6x6x1 |
eV |
RPA_ads_extrap |
RPA adsorbate on slab extrapolated |
eV |
RPA_adsorp |
RPA correlation adsorption energy extrapolated |
eV |
RPA_mol2_ecut425 |
RPA 2nd molecule, Ecut=425 |
eV |
RPA_mol2_ecut450 |
RPA 2nd molecule, Ecut=450 |
eV |
RPA_mol2_ecut475 |
RPA 2nd molecule, Ecut=475 |
eV |
RPA_mol2_ecut500 |
RPA 2nd molecule, Ecut=500 |
eV |
RPA_mol2_ecut530 |
RPA 2nd molecule, Ecut=530 |
eV |
RPA_mol2_ecut560 |
RPA 2nd molecule, Ecut=560 |
eV |
RPA_mol2_ecut600 |
RPA 2nd molecule, Ecut=600 |
eV |
RPA_mol2_extrap |
RPA 2nd molecule extrapolated |
eV |
RPA_mol_ecut425 |
RPA 1st molecule, Ecut=425 |
eV |
RPA_mol_ecut450 |
RPA 1st molecule, Ecut=450 |
eV |
RPA_mol_ecut475 |
RPA 1st molecule, Ecut=475 |
eV |
RPA_mol_ecut500 |
RPA 1st molecule, Ecut=500 |
eV |
RPA_mol_ecut530 |
RPA 1st molecule, Ecut=530 |
eV |
RPA_mol_ecut560 |
RPA 1st molecule, Ecut=560 |
eV |
RPA_mol_ecut600 |
RPA 1st molecule, Ecut=600 |
eV |
RPA_mol_extrap |
RPA 1st molecule extrapolated |
eV |
RPA_slab_ecut300_k12 |
RPA slab, Ecut=300, k=12x12x1 |
eV |
RPA_slab_ecut300_k6 |
RPA slab, Ecut=300, k=6x6x1 |
eV |
RPA_slab_ecut400_k6 |
RPA slab, Ecut=400, k=6x6x1 |
eV |
RPA_slab_ecut500_k6 |
RPA slab, Ecut=500, k=6x6x1 |
eV |
RPA_slab_extrap |
RPA slab extrapolated |
eV |
RPBE_ads |
RPBE adsorbate on slab |
eV |
RPBE_adsorp |
Adsorption energy with RPBE |
eV |
RPBE_mol |
RPBE 1st molecule |
eV |
RPBE_mol2 |
RPBE 2nd molecule |
eV |
RPBE_slab |
RPBE slab |
eV |
adsorbate |
Adsorbate |
|
mBEEF_ads |
mBEEF adsorbate on slab |
eV |
mBEEF_adsorp |
Adsorption energy with mBEEF |
eV |
mBEEF_mol |
mBEEF 1st molecule |
eV |
mBEEF_mol2 |
mBEEF 2nd molecule |
eV |
mBEEF_slab |
mBEEF slab |
eV |
mBEEFvdW_ads |
mBEEF-vdW adsorbate on slab |
eV |
mBEEFvdW_adsorp |
Adsorption energy with mBEEF-vdW |
eV |
mBEEFvdW_mol |
mBEEF-vdW 1st molecule |
eV |
mBEEFvdW_mol2 |
mBEEF-vdW 2nd molecule |
eV |
mBEEFvdW_slab |
mBEEF-vdW slab |
eV |
mol |
Reference molecule 1 |
|
mol2 |
Reference molecule 2 |
|
surf_mat |
Surface Material |
|
vdWDF2_ads |
vdW-DF2 adsorbate on slab |
eV |
vdWDF2_adsorp |
Adsorption energy with vdW-DF2 |
eV |
vdWDF2_mol |
vdW-DF2 1st molecule |
eV |
vdWDF2_mol2 |
vdW-DF2 2nd molecule |
eV |
vdWDF2_slab |
vdW-DF2 slab |
eV |
Similar for the surface energies:
key |
description |
unit |
|---|---|---|
BEEFvdW_bulk |
BEEF-vdW bulk |
eV |
BEEFvdW_slab |
BEEF-vdW slab |
eV |
BEEFvdW_surf |
Surface energy with BEEF-vdW |
eV |
EXX_bulk |
EXX bulk |
eV |
EXX_slab |
EXX slab |
eV |
EXX_surf |
Surface energy with EXX |
eV |
LDA_bulk |
LDA bulk |
eV |
LDA_slab |
LDA slab |
eV |
LDA_surf |
Surface energy with LDA |
eV |
PBE_bulk |
PBE bulk |
eV |
PBE_slab |
PBE slab |
eV |
PBE_surf |
Surface energy with PBE |
eV |
RPA_EXX_surf |
Surface energy with EXX+RPA |
eV |
RPA_bulk_ecut300_k12 |
RPA bulk, Ecut=300, k=12x12x12 |
eV |
RPA_bulk_ecut400_k12 |
RPA bulk, Ecut=400, k=12x12x12 |
eV |
RPA_bulk_ecut500_k12 |
RPA bulk, Ecut=500, k=12x12x12 |
eV |
RPA_bulk_extrap |
RPA bulk extrapolated |
eV |
RPA_slab_ecut300_k12 |
RPA slab, Ecut=300, k=12x12x1 |
eV |
RPA_slab_ecut300_k6 |
RPA slab, Ecut=300, k=6x6x1 |
eV |
RPA_slab_ecut400_k6 |
RPA slab, Ecut=400, k=6x6x1 |
eV |
RPA_slab_ecut500_k6 |
RPA slab, Ecut=500, k=6x6x1 |
eV |
RPA_slab_extrap |
RPA slab extrapolated |
eV |
RPA_surf |
RPA correlation surface energy extrapolated |
eV |
RPBE_bulk |
RPBE bulk |
eV |
RPBE_slab |
RPBE slab |
eV |
RPBE_surf |
Surface energy with RPBE |
eV |
mBEEF_bulk |
mBEEF bulk |
eV |
mBEEF_slab |
mBEEF slab |
eV |
mBEEF_surf |
Surface energy with mBEEF |
eV |
mBEEFvdW_bulk |
mBEEF-vdW bulk |
eV |
mBEEFvdW_slab |
mBEEF-vdW slab |
eV |
mBEEFvdW_surf |
Surface energy with mBEEF-vdW |
eV |
surf_mat |
Material |
|
vdWDF2_bulk |
vdW-DF2 bulk |
eV |
vdWDF2_slab |
vdW-DF2 slab |
eV |
vdWDF2_surf |
Surface energy with vdW-DF2 |
eV |
In the following script it is shown how to extract and plot adsorption and surface energies from the database files (adsorption.db, surfaces.db):
# creates: database_example.svg
import numpy as np
import matplotlib.pyplot as plt
import ase.db
adsorbate = 'NO'
slabs = ['Sc', 'Ti', 'Cu', 'Pd']
db = ase.db.connect('adsorption.db')
db_surf = ase.db.connect('surfaces.db')
labels = ['LDA', 'PBE', 'RPBE', 'BEEF-vdW', 'RPA']
markers = ['o', 's', 'v', 'D', 'x']
markersize = [10, 10, 10, 10, 12]
mews = [1, 1, 1, 1, 4]
cols = ['r', 'k', 'b', 'g']
plt.figure()
for ii, slab in enumerate(slabs):
rows = db.select(adsorbate=adsorbate)
adss = []
for row in rows:
if row.symbols[0] == slab:
adss.append(row.LDA_adsorp)
adss.append(row.PBE_adsorp)
adss.append(row.RPBE_adsorp)
adss.append(row.BEEFvdW_adsorp)
adss.append(row.RPA_EXX_adsorp)
rows_surf = db_surf.select(surf_mat=slab)
surfs = []
for row in rows_surf:
surfs.append(row.LDA_surf)
surfs.append(row.PBE_surf)
surfs.append(row.RPBE_surf)
surfs.append(row.BEEFvdW_surf)
surfs.append(row.RPA_EXX_surf)
for i in range(len(adss)):
if ii == 0:
plt.plot(surfs[i], adss[i], color='darkgray',
marker=markers[i], markersize=markersize[i],
mew=mews[i], label=labels[i])
plt.plot(surfs[i], adss[i], color=cols[ii],
marker=markers[i], markersize=markersize[i], mew=mews[i])
p = np.polyfit(surfs[:-1],
adss[:-1],
deg=1)
plt.plot([surfs[2], surfs[0]],
p[0] * np.array([surfs[2], surfs[0]]) + p[1],
color=cols[ii])
plt.annotate(slabs[ii], (surfs[-1] + 0.02, adss[-1]), color=cols[ii],
size=14)
plt.legend(loc='lower left', numpoints=1, prop={'size': 14})
plt.title('NO adsorption', size=18)
plt.ylim([-3.5, 0])
plt.xlim([0.3, 1.1])
plt.xlabel(r'$E_{\sigma}$ (eV)', size=22)
plt.ylabel(r'$E_{\mathrm{ads}}$ (eV)', size=22)
plt.xticks(size=18)
plt.yticks(size=18)
plt.tight_layout()
plt.savefig('database_example.svg', dpi=500)
Which should generate the following figure showing the adsorption versus surface energy for NO adsorption on four different transition metals:
The surfaces were modeled using three layers with the bottom two layers fixed at the fcc PBE lattice constants from materialsproject.org and the position of the top layer relaxed. The position of the adsorbate was relaxed keeping all three surface layers fixed at the position found previously. All relaxations were carried out with the BFGS algorithm using the PBE approximation to the xc-functional with a force convergence criteria of 0.05 eV/Å. The electron temperature was 0.01 eV and spin-polarized calculations were performed for calculations involving Fe, Ni or Co. 5 Å of vacuum was added to either side of the adsorbate to avoid artificial interactions between neighboring layers following convergence tests at both the DFT and RPA level. The adsorption energies are relative to the molecule in its gas phase and the calculations for the isolated molecules were carried out in a \(6\times6\times6\) Å \(^3\) box fully relaxing the geometry with the PBE functional.
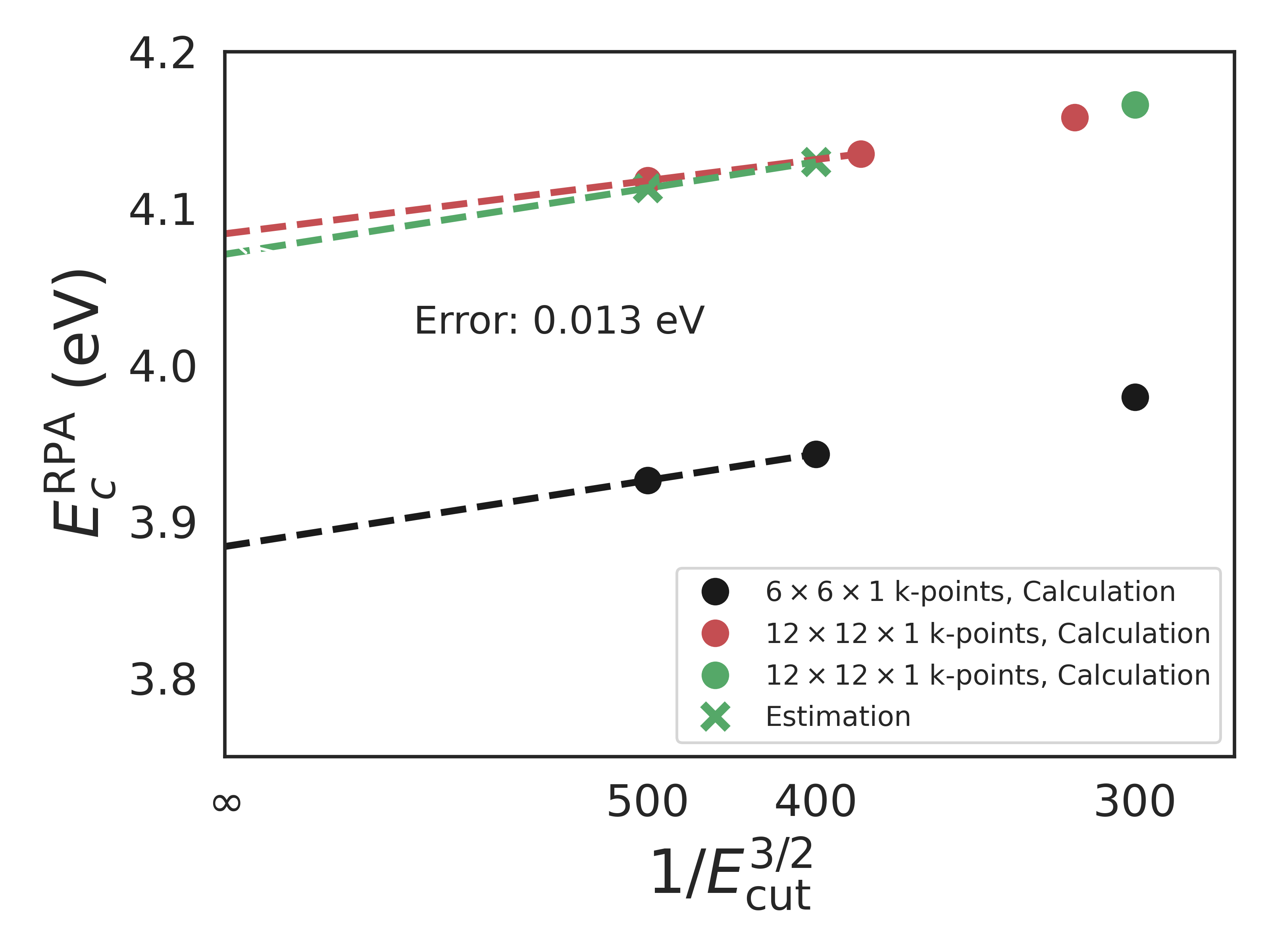
The RPA calculations were carefully converged with respect to plane wave basis using the following extrapolation scheme: In the following figure, the black dots are from a calculation with \(6\times6\times1\) k-points (not enough to achieve convergence) but high cutoff energies (300, 400, 500 eV). The green circle is a calculation at a much denser k-point sampling of \(12\times12\times1\) (converged). From these four circles, the two green crosses are predicted which allow for an extrapolation to infinite cutoff energy. The red dots represent actual calculations with both a dense k-point grid and high cutoff energies to test the extrapolation scheme. The error introduced by the extrapolation scheme for this particular system is seen to be 0.013 eV. The k-point grid of \(12\times12\times1\) ensures that the exchange + correlation energy is converged to within 0.02 eV with respect to the k-point density.